|
 MicrobeTracker saves data in MATLAB format, creating standard .mat files, which
can be opened with MATLAB without running MicrobeTracker. Among other variables,
it saves the variable cellList, the only one that is intended for the
user. To load a saved file in MATLAB, just drag and drop it into the open window
or use the load command.
MicrobeTracker saves data in MATLAB format, creating standard .mat files, which
can be opened with MATLAB without running MicrobeTracker. Among other variables,
it saves the variable cellList, the only one that is intended for the
user. To load a saved file in MATLAB, just drag and drop it into the open window
or use the load command.
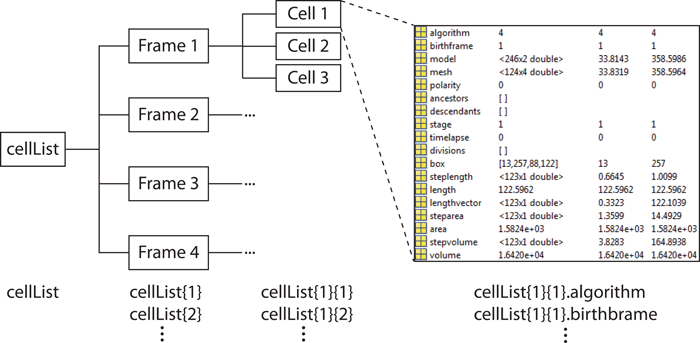
The hierarchical organization of the variable is shown in the image above. It
is a MATLAB cell array, with each cell of the arragy corresponding
to one image frame. Each cell of this cell (subarray) is in turn a cell
array corresponding to a real (that is, biological!) cell. Each of those is a
structure with several fields describing different properties of the
cell. For example, to access the field "algorithm" for the cell 4 on frame 5
type: "cellList{5}{4}.algorithm". The fields of this structure are:
- algorithm - the algorithm used to obtain meshes, see Parameters
section.
- birthframe - the frame on which the cell first appeared in a
timelapse. It is equal to the frame number for independent frames.
- model - the data set describing the shape in MicrobeTracker units, it
is only used by MicrobeTracker to continue detection in timelapses.
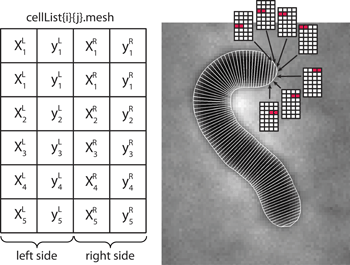
- mesh - the data set describing the shape in geometrical units. This
is a Nx4 matrix (N rows and 4 columns) with the structure shown on the image to
the right. The left two rows of the matrix correspond to the left side of the
cell, for the x and y coordinates respectively. The right two columns
correspond to the right side of the cell. The first row corresponds to the
first pair of points (a cell pole), the second - to the second pair of points
etc. If the cell is aligned (i.e. its polarity is set either manually or
automatically during divisions), then the first row corresponds to the "old
pole", otherwise the vertical orientation of the matrix is random.
- polarity - this property indicates whether the polarity is set
(polarity=1) or not set (polarity=0).
- ancestors/descendents - the list of ancestors or descendents in
genealogical order. If a cell divides, one of the two ("mother") keeps the
number, and the other ("daughter") gets a new one. The daughter keeps the
number of the mother, as well as the grandmother, great-grandmother etc. in
the field ancestors. The mother in turn remember the names of each
daughter in the field descendents. For a more detailed description of
cell numbering see Cell Numbering page.
- divisions - the frame numbers of each division this cell came
through as the mother. The daughter remembers only the frame when it split of
as birthframe.
- timelapse - indicates whether the cell is a part of a time-lapse
series.
- box - the box coordinates (1x4 array with the format: top x, top y,
width, height), that includes the mesh inside. It should be used if cropping
the fragment of the image that contains this particular cell.
- length/area/volume - the length, area, and volume of the cell. The
length is the length of the centerline. The area is the area inside the mesh
outline. The volume is estimated by assuming the height of each segment is the
same as its width and calculated by summing the volume of the cut cones
corresponding to each segment.
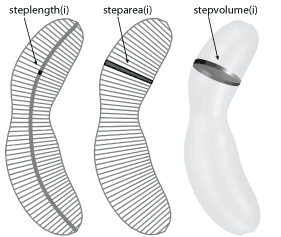
- steplength/steparea/stepvolume - the length, area, and volume of
each segment of the mesh inside the cell. the order is from the old pole to
the new one when the polarity is set, or randomly beginning from one side, if not.
- lengthvector - the coordinates along the centerline of the centers
of each segment. The spacing can be different from the default of 1 pixel and
the polar segments can be different from the set spacing, thus this variable
is more accurate than the number of the row to represent the coordinate.
- signal0/signal1/signal2 - the calculated signal along the cell.
Usually signal0 is reserved for the phase contrast, signal1 for
the images loaded as signal 1 etc.; those these names can be changed (see
Detection and analysis section).
- spots - unlike the rest of the he fields, this one is added by spot
dection tools, sych as a SpotFinderM,
SpotFinderZ, or
peakfinder. This field is a separate
structure that contains information about the location and intensity of
spots detected in a cell. For a detailed description, see the
SpotFinder help page.
|