|
This section describes the interface and various regimes and tools available
in the program. It concentrates primarily on location of the controls and
availability of the tools, while for a sequence of actions required to perform a
particular image analysis task you should refer to
Quick start and
Adjusting parameters sections of
the help.
The tasks that MicrobeTracker performs are organized into several groups
represented by different panels of the main window, which is shown on the image
below. Each subsection describes in detail one of these panels.
Top panel
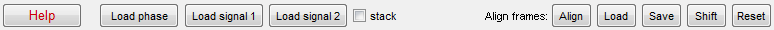
1. Loading images
- Load phase data - loads a set* of phase contrast images (or diffuse
fluorescence images**). By default, MicrobeTracker reads TIFF image
series and will load every image in the selected folder. To exclude one of
the images in a set, simply remove it from the folder. If
BioFormats software
is installed (simply copy loci_tools.jar to MicrobeTracker folder), single
channel stack files produced by several popular microscope control programs
(.stk, .nd2, etc.) can be opened as well. Alternatively, MicrobeTracker will
open multipage TIFF images. To use stacks files or multipage TIFFs, the
checkbox 'stack' has to be selected.
- Load signal 1 / Load signal 2 - loads fluorescence images for
quantification. These images are not used for cell detection; the fluorescence
signal quantification can be done simultaneously during detection or at a
later time following cell detection.
* The program alone works only with TIFF images. The files have to be converted
to TIFF, have the extension .tif or .tiff, and have to be placed into a separate
folder (every TIFF file from this folder will be loaded, any other file will be
ignored). You can easily convert most common formats to TIFF with the help of
ImageJ.
** If fluorescence images (or any other images with light cells on dark
background) are selected, add invertimage parameter
to the list of parameters and set it to 1.
2. Aligning images (for timelapses only)
The alignment procedure (started by the Align button) uses correlation
between successive images in order to compensate for possible microscope stage
or sample drift during image acquisition. The
alignment information is not displayed, and its presence is denoted by active
buttons Save, Shift, and Reset. This information will be
used by the detection routines, but the positions of meshes will be relative to
non-shifted frames. The Shift button will shift the images (phase
contrast and fluorescence) and the meshes and will replace them in memory. This
is designed to review the quality of shifting and to output movies. As a general
rule, it is not recommended to use the Shift button in other cases. The shift
depends on aligndepth parameter, which is 1 by
default and should be increased to 10-20 for slow-drifting image series.
The alignment information can be saved using Save button and loaded
using Load. Reset rezeros the alignment information in memory (but
cannot shift the images back).
Bottom panel
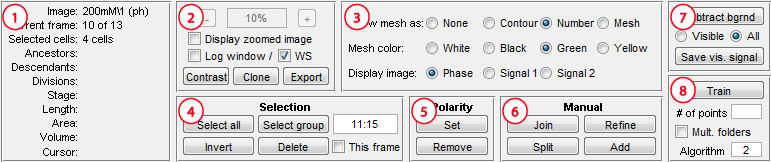
1. Information
This panel is used to display the information about the images
and cells. Image field displays the folder name (only the bottom level)
and the Current frame the number of the image displayed at the moment (as
sorted automatically). The next several fields display the information about the
selected cell or cells, only if at least one cell is selected. All
length-related numeric information is measured in pixels. The last field
displays the cursor position in pixels relative to the image, which can be used
for cropping and outputting images.
2. General image display
Checking the checkbox will display a separate zoomable window.
Its zoom level can be set by the + and - buttons, by the number window
and by the scrolling wheel of the mouse when the cursor is inside the zoomable
window. The next line regulates the visible output, which is always performed to
the Log window (select the checkbox to display) and can be performed to
the MATLAB workspace (select WS checkbox). Contrast regulates image contrast. Clone is the simplest
way of outputting, which replicates the zoomable window in a new window with the menu
that allows saving and coping. Export calls a simple
Image export tool in a separate
window, which allows the user to save the desired region with the meshes and possibly
saves multiple frames and makes movies (only on 32bit systems).
3. Objects to display
This panel allows the user to show/hide meshes, selects their color (black or
white), and selects which image (phase contrast or fluorescence) to display.
4. Selection
Allows the user to select cells. Select all selects all
cells on the frame, Select group selects a group of cells in the
indicated range (the range 11:15 indicated on the figure will select the cells with the
numbers from 11 to 15), Invert inverts the selection. Holding keyboard
Shift button allows for the selecting of multiple cells by clicking on
them. Hold Shift or Control (at the beginning of dragging) and
dragging the cursor to select a group of cells. In this case all cells that
completely fall into the chosen area get selected. Holding Shift inverts
the selection in the area and holding Control deselects all other cells.
The selected cells can be either deleted (using Delete keyboard button or
Delete button on the panel) or used to continue detection in timelapses
for the selected cells only. To undo the last deletion operation, press
Control + Z on the kerboard.
5. Polarity
This group of buttons allows to manually set cell polarity.
Click Set and then label one of the cell's poles as "old pole" (a
little stalk will be displayed and the cell will be marked as having the
polarity set). The polarity can be removed (i.e. considered unknown) if you
press Remove and then click on the cell. Note, the polarity is set
automatically in timelapses when a cell divides.
6. Manual
Use this group of buttons to split, join, or refine selected
cells or add new cells. Additional information on usage of the tools,
particularly the changes in cell numbering and lineage data, is provided on the
Manual Operations page.
- Join - In order to join at least two cells must be selected. The
program will detect the closest poles of these cells and will try to make one
cell by joining the poles.
- Split - The split regime operates in two modes.
The first (semi-autmatic) mode is called by clicking the Split button
and requires exactly one cell to be selected. This mode will determine the
splitting point automatically by finding the minimum "valley" in the phase
image corresponding to the cell.
- Split + Shift - The second mode is called by clicking Split
while holding keyboard Shift button. This mode requires the user to
pick the splitting point. The cell does not need to be selected in advance -
the clicked one will be taken. To exit this mode click outside of the cells of
press Escape. This mode currently operates only when the mesh is
available.
- Refine - In order to refine cells at least one cell must be
selected. The program will run cell detection routine with the current
parameters taking the current cell shape for the starting point.
- Add - Cell addition only works if algorithm 1 or 4 is selected;
in case of algorithm 1 you have to click on the contour, and in case of
algorithm 4 on the centerline (the width is then determined by
cellwidth paramerer); to terminate double click or
press Enter, to cancel press Escape.
- Add+Shift - Hold Shift while finishing cell addition
to call a dialog asking for the new cell number. This number must be a
positive integer and must nor exist on the current or any later frame.
If any of the manual operations fails, no change is made, otherwise the
program replaces the cells in memory, but does not save them to the disk.
Note that the program in these modes also does not automatically integrate
the signal, which has to be added to the meshes separately (for example, by
running it on the selected meshes in 'Reuse meshes' mode, see
below). To undo the last manual operation, press
Control + Z on the kerboard.
7. Background subtraction
Subtracts background from fluorescence images. Meshes
are not used here; the result does not depend on their presence. The "empty"
area used to calculate the background intensity is determined as the light area
on phase images. If Visible option is selected, the background is subtracted
from the fluorescence image set currently being displayed (or none, if phase
contrast images are displayed), otherwise - from all fluorescence images. The
subtracted images can be saved using Save visible signal button.
8. Training
This panel is used to train the algorithm, required for
algorithms 2 and 3 (which use the PDM). The default file is already created for
strain CB15N Caulobacter crescentus cells. The default number of points is 52.
For training you will need an analysis file with a large set of the cells of
correct type, detected using a different method, usually algorithm 1. You can
use multiple files, which will be combined. Enter the number of points (about
the length of the cell in pixels), algorithm (2 or 3), then click Train
and select the data file(s) and the file to save the results.
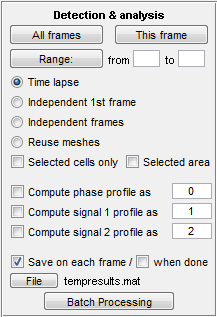
Detection and analysis
This panel is used to perform detection of cells and
quantification of fluorescence. The buttons on top of the panel start the
operation for the selected group of frames: all frames, current frame or a range
of frames. The next set of radio buttons defines whether the images should be
treated as a timelapse (the previous image is used as a guess for the next),
timelapse that starts from a frame number other than one, independent frames
(set of spatially different images), or that the detection should not be
performed at all, only fluorescence image quantification (reuse meshes).
The detection can
also be performed for selected cells only (check Selected cells only). This is
useful in timelapses when different parameters are desired for some cells or
when the detection of all takes too much time. Alternatively it can be performed
for a selected area (check Selected area and select an area holding the Shift
button). The area selection will stay when selecting new frames or performing
analysis on multiple frames. The threshold calculation will only be performed in
this region, therefore this regime can be used to eliminate extremely bright or
dark junk particles in the field of view.
The next block indicates whether the fluorescence signal should
be quantified. By default phase signal will be saved in field
"signal0" (see section "Output format"), 1st fluorescence
signal in field "signal1", the second - "signal2", though
the number can be changed or even replaced with a word or a symbol. This
quantification can be performed while detecting cells or in post-processing,
i.e. when the "Reuse meshes" regime is selected (you can run detection
in the "reuse meshes" regime in order to add the signal to the meshes, which may
be preferable to do at this stage because of larger memory requirements during
cell detection). In any case background subtraction before the operation is
recommended for most applications.
Additionally, you can save the data during processing. By
default, data is saved to the file "tempresults.mat" in the current
MATLAB folder
after each frame. Saving takes time, but allows to recover the data if the
program should crash.
Batch processing
Most operations that MicrobeTracker performs can be automated.
There are two modes designed for this purpose: graphical and text-based.
The graphical mode is a simpler and more widely used mode. It is
called by Batch Processing button, which opens a new window for this purpose,
shown above. Here in each line the user selects where to get files (images,
meshes, parameters). The files currently loaded remain loaded, so you do not
have to reload all of them if you want to keep some of them, for example, the
parameter set. Then you enter which operations you want to perform: currently,
these are background subtraction, cell detection, and signal quantification. To
load images from stack files (if optional Bioformats software is installed),
check the 'stack' box in front of the appropriate button. To remove a selected
file or folder name (to not load it), click on the name itself. At the end of
processing of the whole stack (but not after each frame) the mesh can be saved
into the indicated file. The only specific option for batch processing is "+
prev. frame", which allows to add signal to the frame before processing a range
as well, while identifying cells only within the range. This is designed to pick
"good" cells on the first frame manually and then process the rest of
the frames and add signal to all frames, including the first one.
Lines for the jobs can be added or removed using buttons on the
right bottom side. The batch list can be saved and loaded. It is saved in a text
format, so you can edit the text file directly; this is useful if the jobs are
similar and if there are too many of them. In the latter case you can run the
text file directly, without loading it (there is a limit of 32 jobs if you
try to load or enter them from the interface window).
The text-based batch
mode is called by clicking the
Text mode button. The graphical mode batch window will disappear and will
be replaced with the text one. The window is a simple text editor, which allows
you to type, save and load text files. The files can also be prepared in an
external editor. This window is designed to prepare a simple script, with will
be interpreted by MicrobeTracker when you click Run. The list of
MicrobeTracker-specific commands is shown on top of the window, the interpreter
also accepts MATLAB expressions. This mode was not been rigorously tested yet,
but in principle it allows for a wider range of possibilities than the graphical
mode.
Saving analysis
Saving/loading is straightforward. Click "Save analysis" button
to save the tracking data to a file and "Load analysis" to load it from a file.
The last parameter set used gets saved automatically, but to load it together
with the analysis the checkbox "Load params" needs to be selected. In addition,
there is an option to save only selected cells, for this select "Selection only"
when saving the results.
Parameters
The last control field is designed to enter the
parameters. They
are saved as a simple text file, which can be saved and loaded independently, it
is also saved automatically with the analysis and it can be loaded together with
the analysis. In the analysis file, only the last version of the parameters is
kept. The default parameter set if for algorithm 2. Algorithm 1 requires only a
few parameters and should be changed manually. The default sets for algorithms
2, 3, and 4 (usually the best) are called "alg2.set", "alg3.set", and "alg4.set"
respectively. For the description of individual parameters, see section
"Parameters" below.
Parameter testing mode
Parameter testing mode is
designed to display normally hidden intermediate images for the purposes of
choosing the optimal parameters for a particular image. There are two parameter
testing regimes: Segmentation and Alignment. Segmentation mode lets the user to
preview the segmented image with the current parameter set before doing the
contour alignment, while Alignment mode will display the process of the active
contour fitting to each cell. This mode is only available in algorithms 2-4,
where such alignment is used.
Shift of fluoresence images relative to phase
This regimes allowes to correct for fluorescence images shifted
relative to the phase contrast image used to determine cell outlines. The regime
is called by pressing the keyboard f button. A menu appears, which allows
to set the shifts in x and y direction, which are real numbers measured in
pixels. When you press shift, the meshes in the corresponding fuorescence signal
image will be displayed shifted and will also be shifted when the signal profile
is calculated by MicrobeTracker. Try different shifts manually so that the meshes
look correctly on to of the fluorescence (signal) images. The shift information
can be saved by pressing the r or t (Save As) buttons, and can be
loaded by pressing the y button. This information is also saved with the
meshes, but is not loaded automatically when the meshes are loaded.
Keyboard shortcuts
- Delete - Delete selected cells
- i - Invert selection
- a - Select all cells
- g - Select a group of cells (according to their numbers)
- Up - Go to the previous frame
- Down - Go to the next frame
- s - Save meshes
- l - Load meshes
- Enter - Finish manually adding a cell (in manually adding cell regime)
- Escape - Terminate manually adding a cell (in manually adding cell regime)
- Control+Z - Undo the last manual operation or deletion
- q - Save the list of selected cells to file
- w - Save As the list of selected cells to file
- e - Load the list of selected cells from file
- f - Set the shift of fluorescence images relative to the phase one
- r - Save the shift of fluorescence images to file
- t - Save As the shift of fluorescence images to file
- y - Load the shift of fluorescence images from file
|