|
|
|
joinspots
|
|
A GUI-based program which for linking spots detected with
SpotFinder family tools (SpotFinderZ,
SpotfinderM,
peakfinder). The program can perform automatic
joining with a possibility of manual control and correction.
Usage
Initial setup
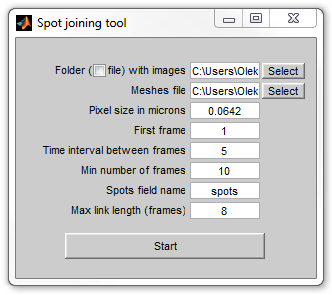
|
|
Initial setup window of joinspots
|
When started, the program opens the initial setup window, where the user
can select the dataset and the parameters of detection. The window will
dissapear when the operation starts, and therefore the parameter can no longer
be changed. The window has the following controls:
-
Folder with images - folder with images that were used for spot
detection to be used for manual control. When the checkbox is selected a
file with images will be opened - either a multipage TIFF or one of the
formats supported by Bioformats when this software is present.
-
Meshes file - a file with meshes and spots produced by SpotFinder
family tools.
-
Pixel size in microns (used as a scaling factor in the output
SpotList arrays, see below).
-
First frame - the first frame where the cells are present. The
program will not consider the cells on the earlier frames.
-
Time interval between frames - will be used for plotting purposes
only.
-
Min number of frames - the program will reject the cells which
existed (were detected) for a shorter period of time.
-
Spots field name - the name of the field that contains spots.
"spots" is the default name used by the SpotFinder family tools, but it can
be changed if multiple colors (e.g. different fluorescence dyes) are used.
-
Max link length - during automatic joining only connect the spots
that are separated by a no longer number of frames.
Main window
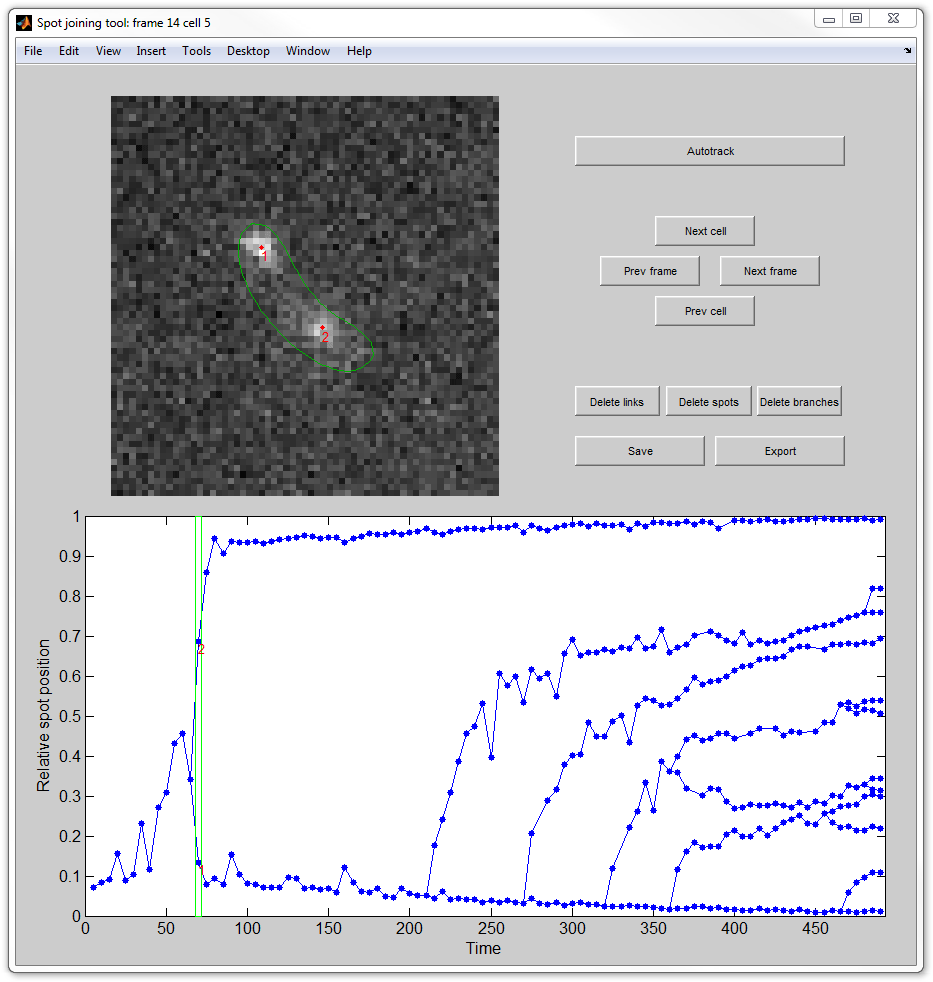
|
|
Main window of joinspots
|
The main window will display an image of the currently selected cell on the
currently selected frame (top left), the tree of spot connections for the
currently selected cell (bottom) with the selected frame highlighted with a box,
and a control panel (top right). The panel contains the following controls:
-
Autotrack - join the spots for the current cell automatically. The
program will forget all the currently available connection and will attampt
to remake them.
-
Next/prev cell/frame - select the next or previous cell or frame.
-
Delete links - when this button is pressed (or keyboard Shift is
held) a click on a spot will delete the link connecting it with its parent.
-
Delete spots - when this button is pressed (or keyboard Alt is held)
a click on a spot will delete the spot and all links connecting it with
other spots.
-
Delete branches - when this button is pressed (or double click is
used) a click on a spot will delete the spot and all its descendants.
-
Save - save the results to a file. The program will prompt for a
file, which will get the three data arrays descrobed below.
-
Export - instead of saving, the output data will appear in MATLAB
workspace.
Output format
The program will output three data arrays:
-
cellList - standard cellList array produced by MicrobeTracker, in
which the field "spots" (or other indicated in the "Spots field name" above)
will get the fields:
-
"parentframe" and "parentnum" - the frame and number of the parent spot;
-
"childrenframe" and "childrennum" - arrays containing the frames and the
number of the children spots.
-
spotList - array of spots, each containing sevaral data fields. Each
of the fields is an array containing the information for every frame on
which the spot existed. The fields are:
-
relpos - relative position of the spot within a cell;
-
abspos - absolute position of the spot within the cell counted from
the old pole;
-
frame - frame number;
-
cell - cell number;
-
spot - spot number within the cell;
-
celllength the length of the cell on that frame;
-
spotList2 - same as spotList, but containing the spots that existed
for at least two frames.
|
|
|